Angel Herráez, PhD. Dep. Biochemistry and Molecular Biology, University of Alcalá. 28871 Alcalá de Henares, Spain
Perceiving molecules as three-dimensional entities is an essential ability to be acquired by students in chemistry and biochemistry. Nowadays plenty of tools are available that allow students to visualize 3D models of chemical structures and even to interact with them on the computer screen using mouse, keyboard and user interface controls like buttons or pull-down menus.
Useful as they are, most of these tools depend on the availability of computer files that contain the atomic coordinates, and optionally other properties, for each molecule under study. These files may or not be readily available, or easy to locate, for an instructor or a student. Hence it is interesting to have a tool that allows anyone to build a model on demand for any envisioned structure. Furthermore, the processes of building one's own structure and conceptually connecting it with the resulting 3D geometry or making small changes and seeing the results adds good educational value to the experience. This method is also amenable to being used for student assignments or tests.
Existing procedures to build models of molecular structures typically require separate software to draw structures and display results, saving of intermediate files, local installation of appropriate software, and learning how to use it. Therefore, although valuable, they hold a limitation for general and widespread use.
With all this in mind, an application has been devised that will allow anyone to draw molecules and build their 3D model easily in a single workspace. We have called it "Do-It-Yourself Molecules".1 This application runs within a web browser without the need for installing special software, does not use a web server and so can be used locally (even from a read-only device like a CD-ROM) as well as through the internet, under most operating systems. It follows a step-by-step approach and brief instructions are provided within the same page, so the learning curve should be smooth and quick.
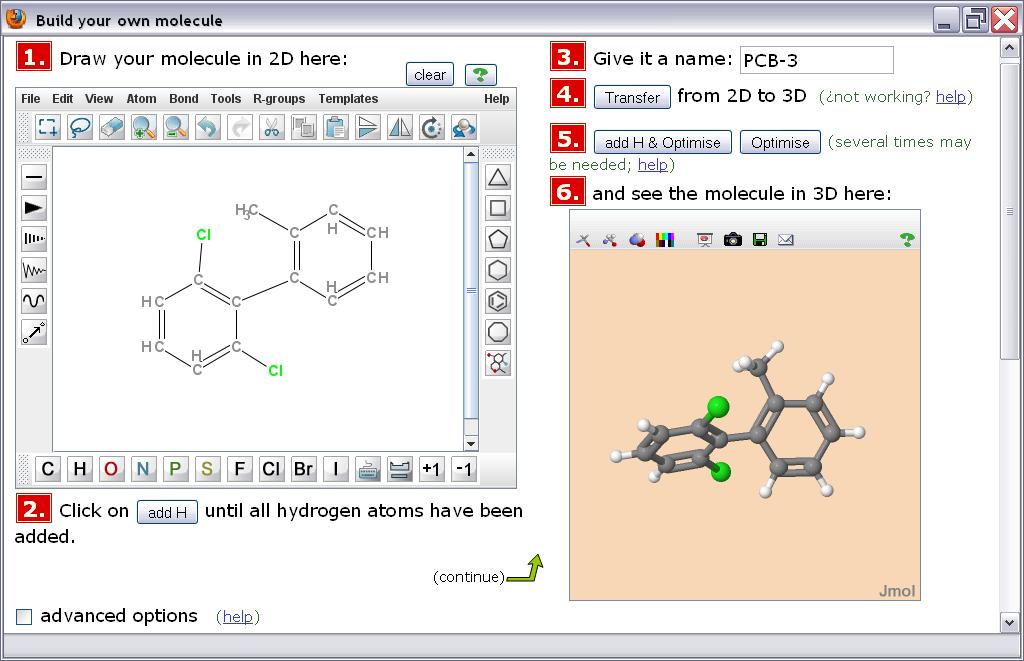
All operations are done in a single window and page within the web browser. The interface integrates numbered steps, a panel for sketching the chemical structure in 2D, a few buttons to drive the generation of an energy-optimized 3D conformation, and a panel where such 3D structure is displayed and available for examination and interaction.
For cases when the structure generated is not what was intended, like a wrong stereochemistry, some tools are implemented to allow a fix or to force guidance in generation of the model. Such "advanced options" will not be needed in most cases and are initially hidden to avoid user confusion or distraction.

Building the application in a webpage provides the advantages of an environment familiar to most users and widespread access without installation, under any operating system that supports Java; it may also be used both online and offline. All software elements involved are available for free and open source. The DIY application itself is offered under a Creative Commons Attribution – Noncommercial – ShareAlike License.
The more polished variant —and currently recommended— of the DIY application uses a JChemPaint applet2 for drawing the structure. This has the advantages of being more powerful, more chemically comprehensive, and localized to several languages. Among other features, it includes an ample set of pre-made structures in the form of "templates", that can be used as such or as a basis for the drawing.
We are also experimenting with alternatives for drawing the 2D structure, like JME, JSDraw, jsMolEdit, SketchEl, Doodler and Kemia.3 Although they are in general less powerful and many are still not fully developed, some offer the advantage of a smaller file size, which accelerates access to the application over slow internet connections. These alternatives may also be tested in the DIY website.
Rendering of the three-dimensional model relies on the Jmol applet.4 This is a powerful piece of software that, apart from display, offers user interactivity, generation of an image, and output of atom coordinates to save or send the resulting model. It is also localized into many languages. The DIY application is currently using the unsigned version of the applet to avoid Java security warnings that may deter users; this imposes the need for some convoluted mechanisms to save results to disk. A more straightforward access policy for file input/output, if so desired, may be achieved by switching to use of the signed Jmol applet.
Most importantly, Jmol is responsible for the computation of the optimized 3D geometry from the 2D sketch. For this, it uses the Universal Force Field (UFF5). Additionally, Jmol can optionally add implicit hydrogen atoms not present in the drawing. Having a full set of hydrogens is essential for a correct 3D optimization. The 2D editor JChemPaint still suffers from a bug that, in some cases, fails to properly add all implicit hydrogens and so this step cannot be automatically implemented in the 2D editor side. Currently, the two options are maintained as user-driven methods: either add the hydrogens in the 2D drawing or add them in the 3D model just before geometry computation.
Connection between the different modules —2D editor, 3D rendering, exported molecular data or image, submission…— is achieved using JavaScript code included in the page source. Particularly, the exchange of molecular data between 2D and 3D panels uses MOLfile6 output and input, handled by JavaScript.
The application also includes a PHP script that will send the resulting 3D model (as text in MOLfile format) to the instructor's email address. For this to work, the DYI application files must reside in a web server that supports the PHP language and sending email with it; this is a requisite fulfilled by commonly used server installations. As an alternative, the student could save the model to local disk and then attach the file to an email message written manually in his/her usual email client.
The interface is simple enough that any occasional user may quickly get acquainted with it and start producing 3D models. It can so be used as support for the instructor during lectures, or be given to students for their own practice or, ideally, for working on assigned exercises. With this is mind, a mechanism has been implemented to allow sending the resulting 3D model to the instructor by email.
The process of drawing the flat structural formula and seeing it immediately converted into a model in 3 dimensions may by itself have enough formative value for students. Furthermore, the system offers easy chances to play and test variations of the structure initially planned. Apart from this, the optimization of the flat structure towards a 3D conformation is displayed in real time, adding great pedagogical value, as the user sees the bonds stretching and wiggling until the model reaches a proper stereochemistry. Along the same line, one of the advanced options allows to pull atoms with the mouse and see immediately how they struggle to fit back into proper positions compatible with the stereochemistry —but not necessarily the same they had initially—.
In addition to the basic functionality, some utilities are included. The displayed 3D model can be duplicated in an enlarged window (that can be resized up to full screen), useful e.g. for detailed inspection of the structure or for projection in a classroom. Also, a still image or snapshot of the 3D model can be captured and then it may be copied or saved to file.
The DIY application is freely available for use.1 The interface is offered in either English or Spanish; translation into other languages could be easily arranged. Installation of a copy on a server, or even locally in a computer room, is straightforward (one just needs to copy several files, under 9 MB in total). All the software used is open source and cross-platform, and the application is offered under a Creative Commons license (Attribution – Noncommercial – ShareAlike 3.0).
1. Do-It-Yourself Molecules: from 2D to 3D. A. Herráez (2010) http://biomodel.uah.es/en/DIY/ [accessed 17 Oct. 2010]
2. JChemPaint: an open-source editor for 2D chemical structures, http://sourceforge.net/apps/mediawiki/cdk/index.php?title=JChemPaint
3. JME Molecular Editor, http://www.molinspiration.com/jme/
JSDraw, http://www.chemene.com:8080/ccd/jsdraw/
jsMolEdit, http://chemhack.com/jsmoleditor/component/content/frontpage
SketchEl, http://sketchel.sourceforge.net/
Doodler, http://web.chemdoodle.com/
Kemia, http://kemia.github.com/
4. Jmol: an open-source Java viewer for chemical structures in 3D, http://www.jmol.org/
5. UFF, a rule-based full periodic table force field for molecular mechanics and molecular dynamics simulations. A. K. Rappé, C. J. Casewit, K. S. Colwell, W. A. Goddard III, and W. M. Skiff (1992) J. Am. Chem. Soc. 114, 10024-10035. doi:10.1021/ja00051a040
See also:
http://web.archive.org/web/20060110100545/http://franklin.chm.colostate.edu/mmac/uff.html [accessed 17 Oct. 2010]
http://towhee.sourceforge.net/forcefields/uff.html [accessed 17 Oct. 2010]
6. The MOLfile, originally from MDL, is a widespread file format for 2D and 3D molecular structures compatible with many software programs. http://www.symyx.com/solutions/white_papers/ctfile_formats.jsp