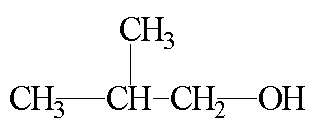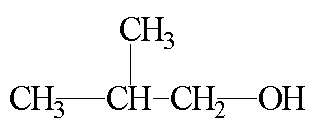Spectrum in GIF format (5 k)
Coupling and NOE in Carbon-13 NMR
This set of spectra show the effect of carbon-proton coupling (splitting of carbon
peaks) and NOE on the C-13 spectrum.
The coupled carbon spectra are useful for verifying chemical shift assignments.
The CH3 carbons at 19 ppm are split into a quartet, the CH carbon
at 31 ppm is split into a doublet, and the CH2 carbon at 70 ppm is
split into a triplet. The coupling constant for these splittings, approximately 125 Hz,
is consistant with C-H coupling and the splitting patterns are consistant with the
previous shift assignment.
The effect of the NOE is apparent when the different pulse sequences
are compared. The CDCl3 triplet at 77 ppm. is not effected by the
proton decoupling because the deuterium signal is at a different frequency. As a result
it is a useful internal standard when comparing experiments.
Decoupled C-13 Spectrum. This is a typical C-13 spectrum with
broadband decoupling of protons.
FID in NUTS format (263 k)
Spectrum in Replica format (35 k)
Spectrum in GIF format (4 k)
Coupled C-13 Spectrum. This is a C-13 spectrum with the decoupler
turned off to produce a coupled C-13 spectrum.
FID in NUTS format (263 k)
Spectrum in Replica format (48 k)
Spectrum in GIF format (4 k)
Decoupled C-13 Spectrum without NOE. In this experiment the decoupler
is only on during data aquisition. This produces a decoupled spectrum
but does not allow NOE to build up. The effect of the NOE is apparent when
you compare the intensity of these peaks with the regular decoupled C-13
spectrum.
FID in NUTS format (263 k)
Spectrum in Replica format (30 k)
Spectrum in GIF format (4 k)
Coupled C-13 Spectrum with NOE. In this experiment the decoupler is
off during aquisition, but it is turned on between aquisitions. This produces a
coupled C-13 spectrum, but the NOE has time to build up between acquisitions.
The S/N improvement from the NOE is apparent when this spectrum is compared
with the regular coupled C-13 spectrum.
FID in NUTS format (263 k)
Spectrum in Replica format (48 k)
Spectrum in GIF format (5 k)
Spectral Editing (APT and DEPT)
This set of spectra show how special pulse techniques like DEPT and APT can
help interpret NMR spectra. The information from these experiments
is very similar to that obtained from a coupled C-13 specturm.
APT spectrum. This pulse sequence produces a C-13 spectrum where
C and CH2 carbons are inverted relative to CH and CH3
carbons.
FID in NUTS format (263 k)
Spectrum in Replica format (64 k)
Spectrum in GIF format (5 k)
DEPT-45. This pulse sequence produces a spectrum where all carbons
are observed. The polarization transfer from the pulse sequence
significantly increases the S/N for carbons with attached protons.
FID in NUTS format (132 k)
Spectrum in Replica format (32 k)
Spectrum in GIF format (4 k)
DEPT-90. This pulse sequence produces a spectrum where only CH carbons
are observed (Small peaks are observed for other carbons IF the delays
are not just right in the pulse sequence).
FID in NUTS format (132 k)
Spectrum in Replica format (40 k)
Spectrum in GIF format (4 k)
DEPT-135. This pulse sequence produces a spectrum where the peaks for
CH2 carbons are inverted, CH and CH3 carbons are up.
FID in NUTS format (132 k)
Spectrum in Replica format (31 k)
Spectrum in GIF format (4 k)
T1 Inversion Recovery Data
The T1 inversion recovery experiment is used to determine
the time constant for spin lattice relaxation. This is useful for
studying physical processes and of practical importance for
preventing saturation during NMR experiments. This data set is available
as a 2D NUTS file containing processed spectra or as a stacked plot in
*.gif and *.rpl format. NUTS includes macros to process
this T1 data (see the help menus in NUTS). A Lotus 123 v4
worksheet with the data workup is included for the proton T1
experiment.
Proton T1 Inversion Recovery
2D spectrum NUTS format
Stacked plot in Replica format
Stacked plot in GIF format
Lotus worksheet
Carbon-13 T1 Inversion Recovery
2D spectrum NUTS format
Stacked plot in Replica format
Stacked plot in GIF format
2-D NMR Data
- COSY. This experiment shows Proton Proton correlation through scalar (through bond) coupling. The same coupling that causes splitting. Both axes are proton 1-D spectra. The matrix diagonal also shows the 1-D spectrum. The off-diagonal peaks show which protons are correlated.
- HETCOR. This experiment HETeronuclear CORrelation between Proton and Carbon nuclei with scalar coupling. One axis is the 1-D proton spectrum, the other axis is the 1-D carbon spectrum. Observed peaks show correlation between protons and attached carbons.
Return to NMR Data at Widener University.
This page is maintained by Scott Van Bramer
Please send any comments, corrections, or suggetions to
svanbram@science.widener.edu.
This page has been accessed
times since 1/5 /96 .
Return to Scott Van Bramer's Home Page
Widener University Science Division Home Page
Last Updated 1/5/96